ATLAS¶
Quick Start¶
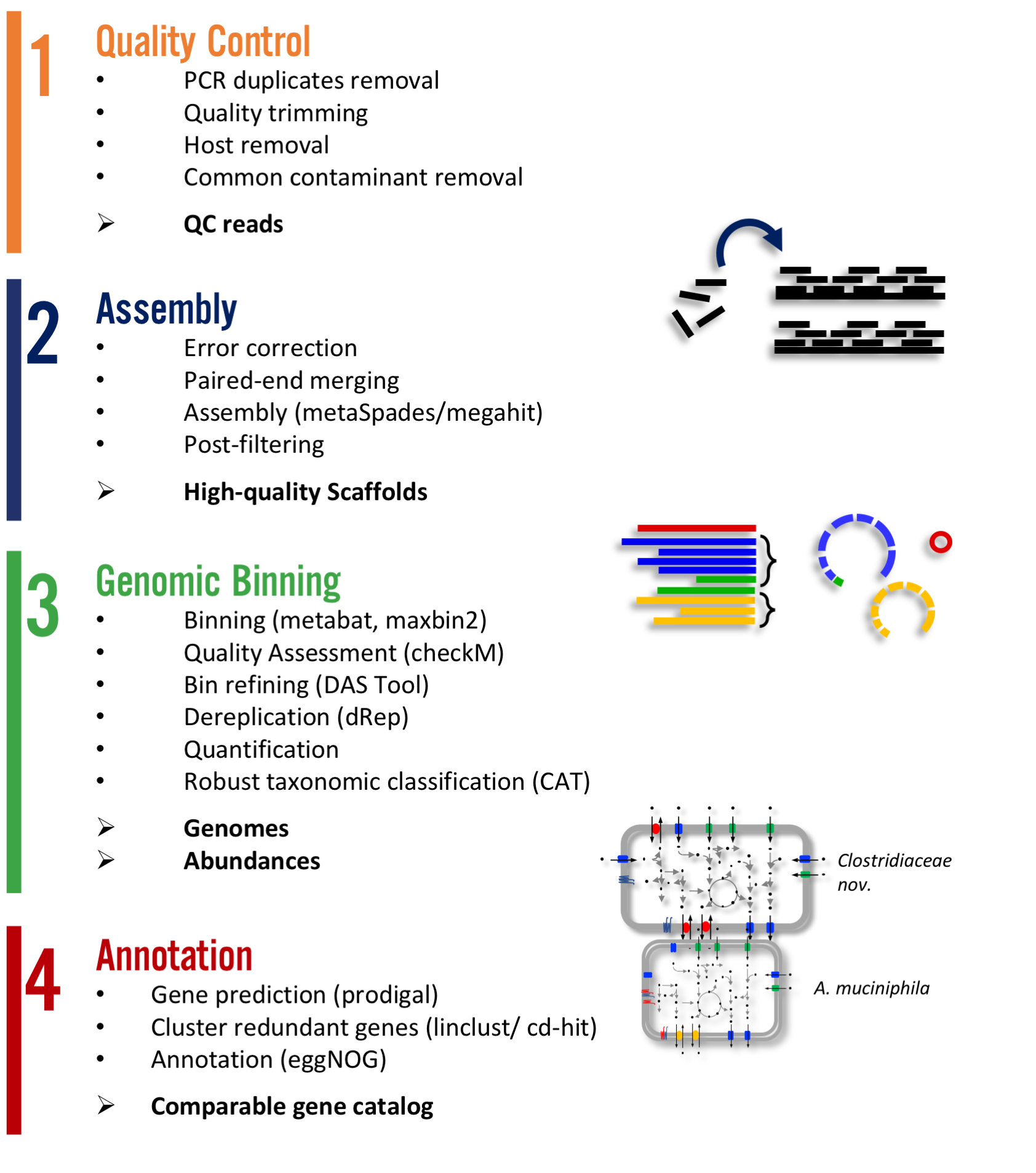
Three commands to start analysing your metagenome data:
conda install -c bioconda -c conda-forge metagenome-atlas
atlas init --db-dir databases path/to/fastq/files
atlas run
All databases and dependencies are installed on the fly in the directory --db-dir.
You want to run this three commands on the example_data on the GitHub repo.
If you have more time, then we recommend you to configure atlas according to your needs:
- check the samples.tsv
- edit the config.yaml
- run atlas on a cluster system